Research Interest
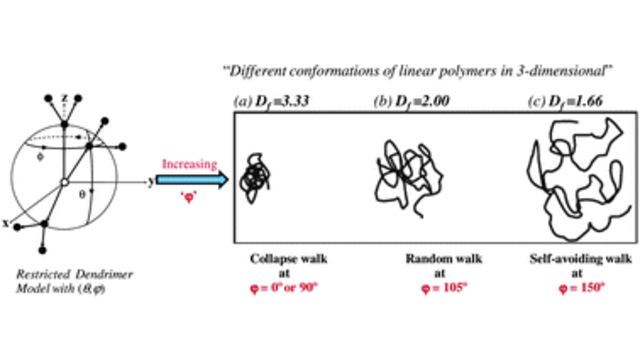
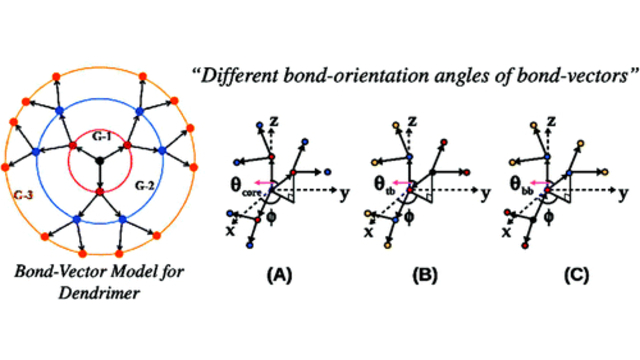
Dendritic Polymers: The conformational and dynamic properties of dendrimers and randomly hyperbranched polymers are explored in the framework of optimized Rouse-Zimm models as a function of flexibility and excluded volume parameters and generational growth.
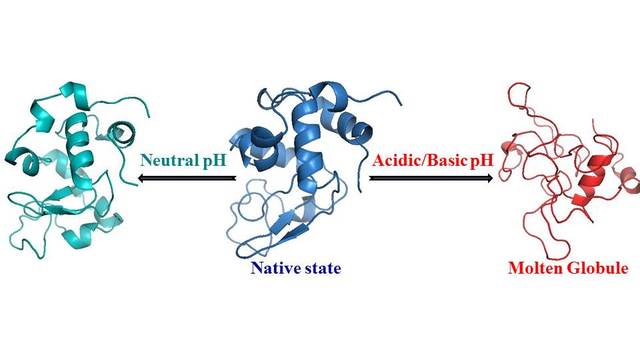
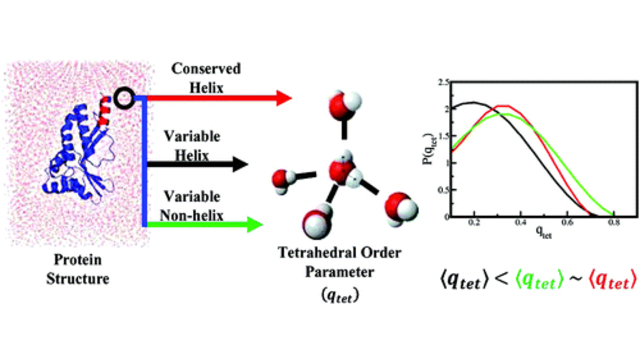
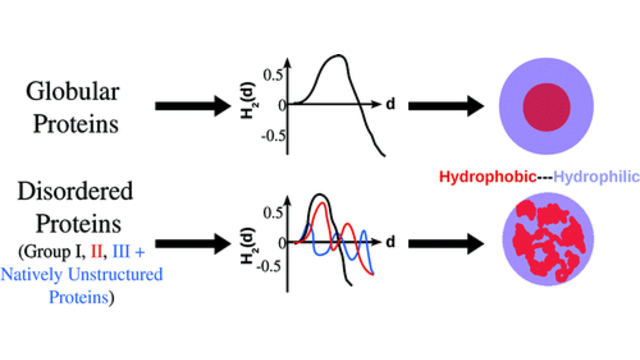
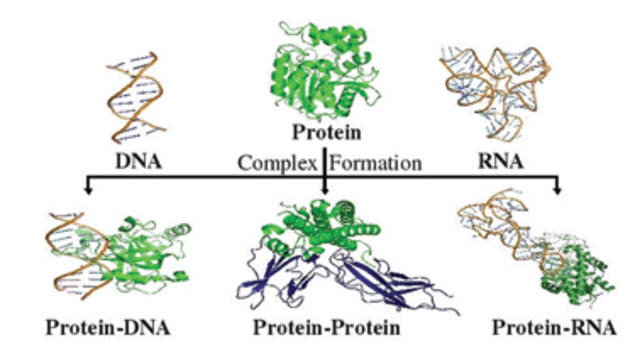
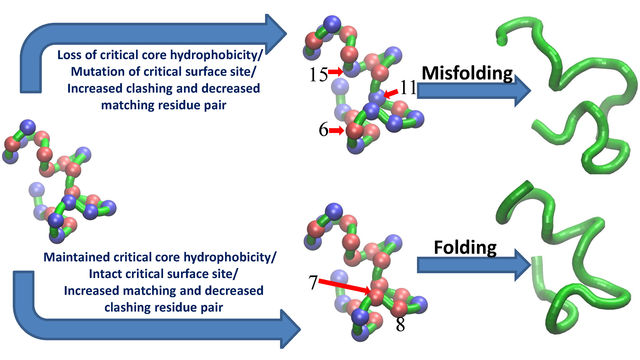
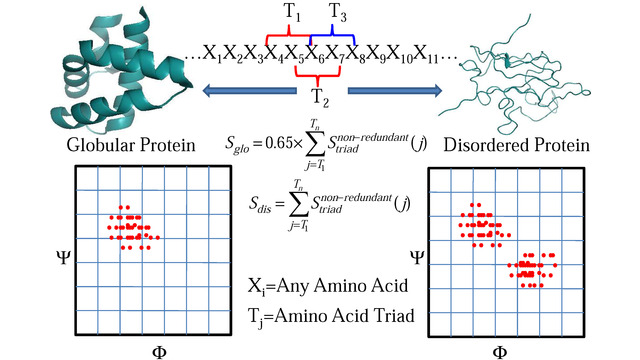
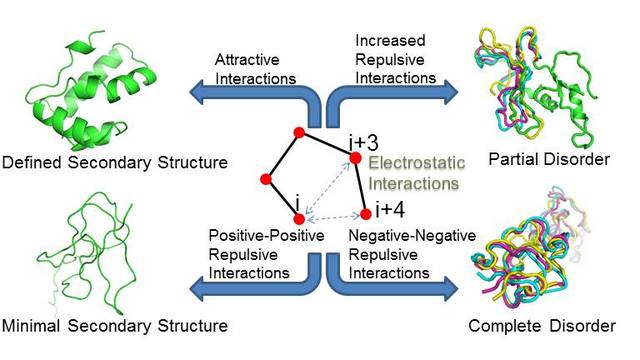
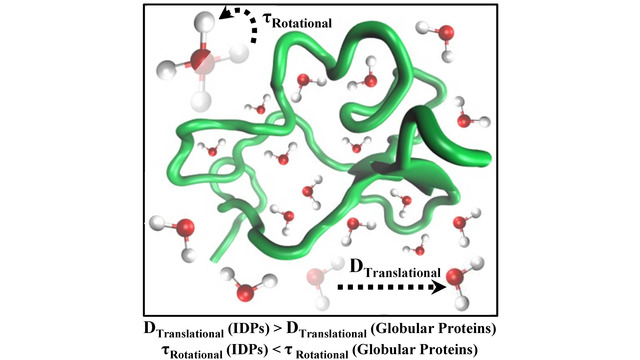
more...Proteins: State-of-the-art computational and theoretical techniques are used to study the structure and dynamics of protein and its environment. A major goal is to understand protein misfolding, intrinsic disorder and fold switch through sequence design and sequence analysis.
more...Recent Publications
1. S. Karmakar and P. Biswas, 2026, Conformations of polyglutamate chains near single walled carbon nanotubes, Biophysical Chemistry, 331, 107581
1. S. Kumar and P. Biswas, 2026, Conformations of Semiflexible Ring Polymers, Phys. Chem. Chem. Phys., 28, 2652
2. M. Handa and P. Biswas, 2026, Conformations and dynamics of ring polymers in dilute solutions, Polymer, 344, 129485
3. S. Karmakar and P. Biswas, 2025, Local Structure and Dynamics of Hydration Water in Amyloid-β Aggregation and Caffeine-Mediated Inhibition, J. Phys. Chem. B, 129, 8858
4. R. Singh and P. Biswas, 2025, A physical model of Fourier uncertainty principle, Europhys. Lett., 151, 10001
5. R. Singh and P. Biswas, 2025, Evaluating multi-state free energy profiles from splitting probability, J. Chem. Phys., 162, 055102
6. S. Kumar and P. Biswas, 2024, Rheology of Ring Copolymers in Dilute Solutions, J. Phys. Chem. B, 129, 496
more...







 1
1 2
2 3
3 4
4 5
5 6
6 7
7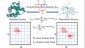 8
8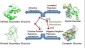 9
9 10
10 11
11 12
12 13
13 14
14 15
15 16
16